

The extreme C-terminus contains an EEVD motif that interacts with cochaperones that possess degenerate 34 amino acid tetratricopeptide repeats (TPRs). Hsp70 function and specificity are highly regulated by cochaperones such as Hsp40s and nucleotide exchange factors (NEFs) (described below).Īll Hsp70s are composed of three domains, an ~44 kDa N-terminal ATPase domain, an ~15 kDa substrate binding domain, and an ~10 kDa C-terminal lid. Hsp70s are highly conserved and are found in all organisms. Indeed, Hsp70s are perhaps the best-studied class of chaperones.
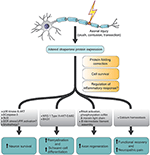
Members of the Hsp70/Hsc70 class of molecular chaperones are essential for protein folding, membrane translocation, disrupting protein aggregates, and rearranging multiprotein complexes. If, however, this load cannot be decreased, ATF4 and ATF6 dependent apoptosis may ensue. Along with activation of Ire1, these cascades reduce the overall protein load in the mammalian secretory pathway. In mammals, the ER sensor, ATF6, upregulates the synthesis of ER chaperones, and the ER transmembrane kinase PERK activates the synthesis of the ATF4 transcription factor but triggers a general inhibition of translation. Hac1 regulates the transcription of UPR targets that include chaperones and ERAD effectors. In yeast, misfolded proteins in the ER are sensed by the transmembrane kinase, Ire1, which when activated promotes the splicing of the message for the Hac1 transcription factor. In most cases, the accumulation of misfolded proteins in the ER leads to the activation of the unfolded protein response (UPR). For example, cystic fibrosis, antitrypsin deficiency, Alzheimer’s disease, Huntington’s Disease, and nephrogenic diabetes insipidus result when specific proteins either fail to fold properly and are targeted for degradation, or are hidden from the degradation machinery and accumulate as toxic aggregates. Not surprisingly, many diseases are linked to defects in the ER associated quality control machinery (ERQC). In fact, all three chaperone classes are involved in targeting proteins for degradation in addition to catalyzing folding. The same chaperones involved in “folding” the substrate may also trigger its proteolysis. For example, a different set of chaperones is responsible for recognizing the defect and targeting the protein for degradation depending upon where the folding lesion is located within the protein. Because transmembrane proteins have domains exposed in the ER lumen, in the cytosol, and in the membrane, this group of proteins may interact with factors in each of these compartments. The thiooxidoreductase proteins mediate proper disulfide bond formation and may also act as chaperones. Similarly, the ER lectins bind to immature glycoproteins to promote substrate folding and will retain proteins in the ER in the event folding is compromised. Several ER lumenal heat shock protein homologues prevent protein aggregation and promote folding. Three major groups of ER chaperones act on the nascent chain, heat shock protein homologues (e.g., Hsp70, Hsp40, Hsp90), the ER lectins (e.g., calnexin, calreticulin), and the thiol oxidoreductases (e.g., PDI).
Next, as translocating proteins leave the translocon, folding and post-translational modifications are mediated by a collection of chaperones and enzymes. These three events: protein translocation, protein folding, and protein degradation, are each facilitated by a group of factors termed molecular chaperones.īefore the ribosome-nascent chain complex even docks at the ER, molecular chaperones play an important role in ER physiology: The ER lumenal Hsp70 chaperone, BiP, maintains the permeability barrier between the cytosol and the ER lumen by gating the Sec61 translocon. Next, the protein will fold properly and traffic to its final destination, or if folding is inefficient the protein can be targeted for ER associated degradation (ERAD) by the cytosolic proteasome. Proteins that contain an Asn-X-Ser/Thr sequence are glycosylated by oligosacchhryl transferase as the polypeptide emerges from the translocon. Soluble proteins are translocated into the ER lumen, whereas transmembrane proteins are cotranslationally integrated into the lipid bilayer. After translation is initiated in the cytosol, the ribosome-nascent chain complex docks at the Sec61 translocon in the ER membrane and protein synthesis resumes. Approximately 30% of all newly synthesized proteins are delivered to their final cellular destinations via this pathway. The endoplasmic reticulum (ER) is the entry point for proteins in the secretory pathway. Introduction: Protein Biogenesis in the ER


 0 kommentar(er)
0 kommentar(er)
